More Information
Submitted: December 20, 2021 | Approved: January 11, 2022 | Published: January 12, 2022
How to cite this article: Kumar N, Jain R, Seerat, Chauhan A, Verma D. Rapid and sensitive identification of cow and buffalo species and gender in tissue/meat samples impounded from different spots in Delhi NCR India by Real Time PCR. J Forensic Sci Res. 2022; 6: 012-016.
DOI: 10.29328/journal.jfsr.1001031
Copyright License: © 2022 Kumar N, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Real time PCR; Adulteration; Species identification; mtDNA; DNA testing; Meat labeling
Rapid and sensitive identification of cow and buffalo species and gender in tissue/meat samples impounded from different spots in Delhi NCR India by Real Time PCR
Naresh Kumar1*, Rahul Jain1, Seerat1, Amit Chauhan2 and Deepa Verma1
1Forensic Science Laboratory, Home Department, GNCT OF Delhi, Rohini, Delhi, India
2Department of Life Science, School of Sciences, Christ University, Bengaluru-560029, India
*Address for Correspondence: Naresh Kumar, Forensic Science Laboratory, Home Department, GNCT OF Delhi, Rohini, Delhi, India, Email: [email protected]
The objective of this study was to obtain a fast, accurate and reliable method of species identification of unknown biological samples for forensic applications, especially in illegal trade of animals as well as meat fraud. Meat fraud and adulteration not only affects the market but also increases the risk of religious and ethnic conflicts around the world [1]. In this study, species-specific and gender differentiating Real time PCR technique was employed to analyse 15 meat samples collected from a suspected site. Out of 15 samples collected from suspected site, 54% and 13% samples were of Cow and buffalo origin respectively. All 54% cow samples were of male while one each of buffalo were of male and female origin. Two samples were inconclusive. These findings indicated that species and gender-specific PCR is very sensitive and can be used for forensic species identification and the detection of meat fraud and adulteration.
India is one of the world’s largest meat producers and ranks eighth in the world’s meat production. There are 1176 APEDA (Agricultural & Processed Food Products Export Development Authority) registered slaughterhouses and 109 meat processing plants, however, illegal slaughterhouses contribute major challenges in the authenticity of the meat produced. Even though cow slaughter and transport of cow meat is banned in most of the Indian states, smuggling of cow and its meat nationally and internationally has been reported [2]. Slaughtering of buffalo and goat in illegal slaughterhouses is also prohibited and comes under the preservation of Cruelty to Animals Act.1960.
Authentication of the meat species is important due to problem of meat adulteration or substitution of meat with that of cheaper or less acceptable quality. Besides consumer satisfaction, certain social and religious concerns and possible health hazards associated with particular type of meat warrants honest labeling of the source of meat and meat products. Additionally, believers of Hinduism and Islam are averse from inclusion of beef and pork in their diet, respectively, owing to their religious beliefs [3]. Some of the reasons which necessitate species and sex identification are :(a) prevention of economic fraudulence of misrepresenting costlier meat with cheaper meats; (b) implementation of statutory slaughter restrictions on cattle especially cow; (c) verification of export consignments [4,5].
Traditional morphological analysis or other serology-based analyses are usually applied to identify undeclared meats, but these methods often produce incorrect or unreliable results [6]. Therefore, using more precise and reliable method is critical in food authentication.
For past several decades technologies based on the DNA analysis have become more sensitive and reliable for the identification of species in row and processed meat [7], and from difficult to process samples like hair, and bone as well [8]. Various DNA genes of mitochondria are used as the target for detection and identification of meats from animal species [9]. It has been observed that Mitochondrial DNA has advantage over nuclear DNA when using it as a source of species specificity as it has remained unaffected during the course of evolution and contains several conserved genes. Mt DNA is present in thousands of copies per cell which helps in identification of species even for poor quality of samples. Since Mt DNA follows maternal inheritance, it is free of heterozygosity [10] and allows the discrimination of even closely related species. Cyt b gene is one of the genes which is conserved [3,11] and AMELogenein (AMEL) a gene present on sex chromosomes (X and Y) was used for sex determination [12] in present study.
Real time duplex PCR was used to identify and differentiate cow and buffalo meat and determination of sex using Mylab discovery solutions kit from suspected meat samples received from various crime spots in the study. This work presents a specific, sensitive, effective, and fast method for reporting adulteration of meat and meat products along with gender identification.
Sample collection and DNA extraction
A total of 15 meat samples collected from a suspected site and submitted for evaluation by Delhi police was used in this study.
All kits used in this study were supplied by Mylab discovery solutions Pvt. Ltd. Pune.
DNA was isolated from meat samples using Maverick nucleic acid extraction kit (Mylab discovery solutions, Pune, India) as per the manufacturer’s specifications. Each 25 mg of meat sample was separately minced and washed with 160 µl of 1 X Phosphate buffer saline (Himedia). DNA was eluted in 100 µl of Elution buffer and stores at 4 °C till further downstream process.
Real-time PCR with the commercial kit
The Cow species identification kit, Buffalo species identification kit and cattle sex determination kit (Mylab discovery solutions, Pune, India) were used to perform real-time PCR. The assay is supplied with positive control (PC) and internal control (IC), allowing for monitoring of PCR inhibition. The reactions were carried out on CFX-96 real time PCR system (BIO-RAD) with a final reaction volume of 20 μl. All the samples were tested with following conditions: UNG incubation at 50 °C for 2 min followed by polymerase activation for 10 min at 95 0C and 45 cycles of denaturation at 94 °C for 15 seconds and data collection at 60 °C for 45 seconds. The samples were determined to be positive for respective assay if the Ct value was ≤ 37.
Determination of cow and Buffalo species-specific mtDNA using Real Time PCR
Recently, several issues related to meat adulteration arose in many countries around the globe. The impact of the adulterated meat scandal was higher in countries with communities for whom meat consumption pattern is driven by cultural preferences and religious beliefs. Detection of undisclosed animal species in meat products is important not only for protecting the consumer from fraud but also to guarantee the respect of his religious beliefs and cultural preferences [3]. To assess the status of meat labelling and species authenticity, a variety of DNA-based testing methods are employed. Earlier methods were based on identification of proteins by means of electrophoretic and/or immunological methods [13,14]. However, these techniques are not reliable for identifying species in poorly stored samples and or complex heated products because of deterioration of the proteins. DNA is more stable than proteins during heat processing and although it can be fragmented by heat, modern DNA techniques still allow us to identify DNA from the different species present in a sample [10,15]. Different DNA-based techniques have been used for meat authentication, which include DNA hybridization [16]; PCR-RFLP [17-19]; RAPD-PCR [20]; PCR-SSCP [21] Among these techniques, Real time PCR is considered as a highly discriminatory and reliable [22]. This technique is particularly suitable because even small fragments of DNA formed during processing of the meat can still be amplified and identified [23]. PCR based assay developed for species identification commonly utilized Mitochondrial DNA (mtDNA) sequences [24].
In this study, the primary objective was to screen meat collected from crime spots in Delhi NCR region for adulteration using an accurate, robust, reliable method. We opted to use real-time PCR and specific primers/probe set within highly sensitive system such as VetScreenTM Cow Species Identification kit for the identification of cow species and VetScreenTM Buffalo Species Identification kit for identification of Buffalo species using mitochondrial Cytb gene to identify meat samples while VetScreenTM Cattle Sex Determination kit was used for discrimination of sex in submitted samples. (Manufacturer: Mylab discovery solutions Pvt Ltd Pune). The quality of DNA is an important factor to consider in the design of DNA analysis for food authentication. In our study, the Mavrick DNA extraction kit (Mylab discovery solutions Pvt Ltd Pune) provided an excellent quality and concentration of DNA preparations.
We performed cow and Buffalo species identification and sex determination by Real Time PCR simultaneously for all 15 meat samples collected from different crime spots by Delhi police. For both Cow and Buffalo identification, kit includes an endogenous internal control to monitor extraction and PCR inhibition during the process.
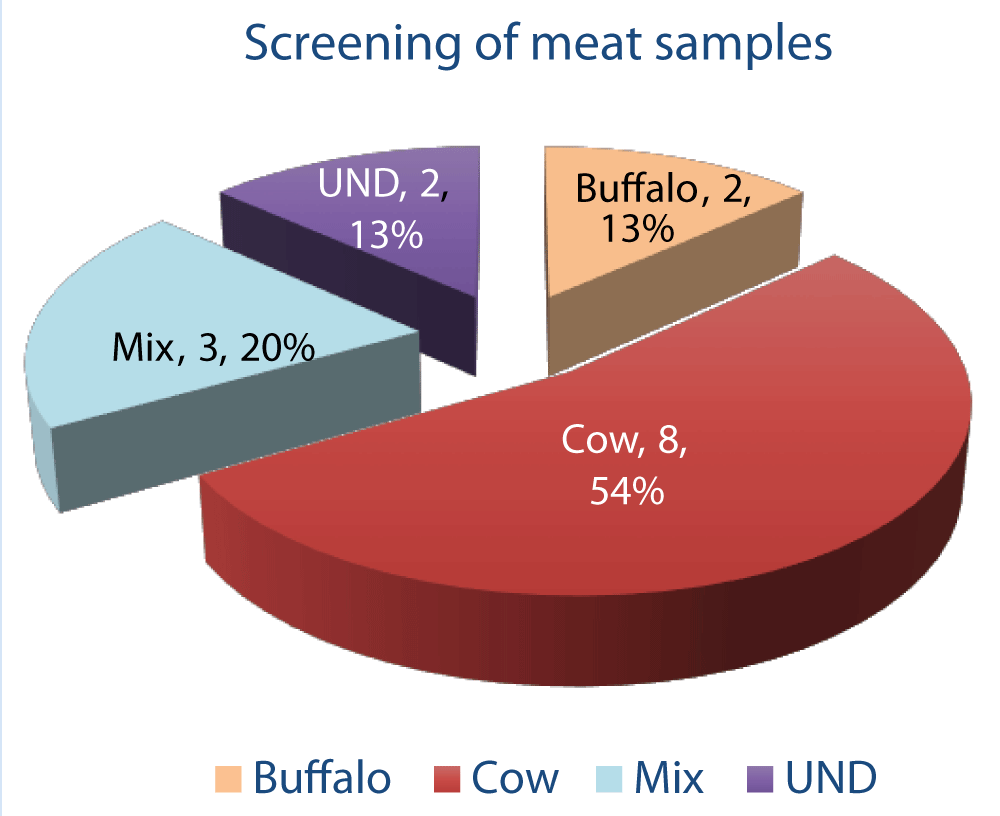
Out of 15 samples submitted for identification, 13% (N = 2) were of buffalo (Table 1) and 54% (N = 8) were of beef origin (Table 2). 20% (N = 3) samples were mix of beef and buffalo meat (Table 3) while 13% (N = 2) samples out of 15 could not be identified (Figure 1).
| Table 1: Buffalo. | |||
| Sample No | Biological Set Name | CqFam | Cq Vic |
| Sample 1 | buff | 17.30 | 40.36 |
| Sample 2 | buff | 19.93 | 30.46 |
| Table 2: Cow. | |||
| Sample No | Biological Set Name | Cq Fam | Cq Vic |
| Sample 6 | cow | 21.73 | 26.78 |
| Sample 7 | cow | 25.92 | 31.13 |
| Sample 8 | cow | 25.41 | 26.39 |
| Sample 9 | cow | 28.85 | 32.59 |
| Sample 11 | cow | 27.80 | 31.07 |
| Sample 13 | cow | 13.40 | 30.97 |
| Sample 14 | cow | 17.35 | 35.72 |
| Sample 15 | cow | 14.87 | 35.20 |
| Table 3: Mix meat. | ||||||
| Sample No | Biological Set Name | CqFam | Cq Vic | Biological Set Name | CqFam | Cq Vic |
| Sample 4 | buff | 34.91 | 32.69 | cow | 24.46 | 29.71 |
| Sample 5 | buff | 31.16 | 33.97 | cow | 28.57 | 33.06 |
| Sample 10 | buff | 39.89 | 31.02 | cow | 26.29 | 30.67 |
Figure 1:
Gender determination
In last few decades sensitive and reliable PCR based methods targeting genes present on sex chromosomes (X and Y) e.g. AMELogenin locus-AMELX and AMELY [25], sex-determining region of the Y chromosome (SRY) gene [26] have been developed. European union have the special guidelines and rules (The Commission Regulation (EC) 765/2002) that recommend the use of PCR based methods for sex identification to export male beef meat [27].
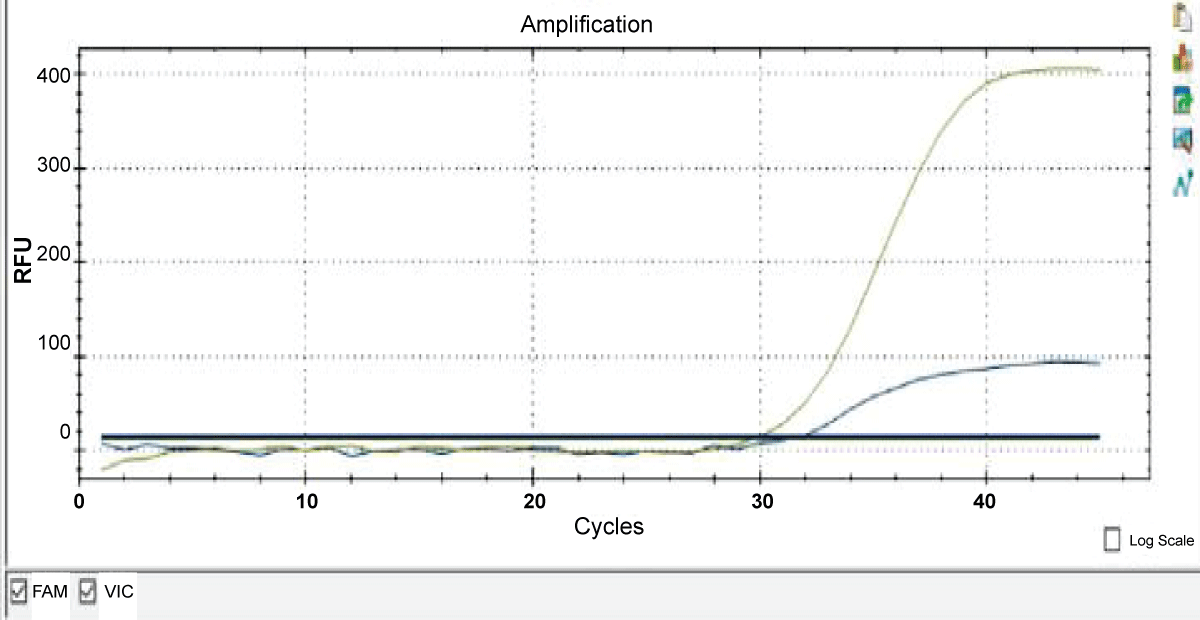
Gender identification was carried out for all 15 samples which were tested for species identification. Mylab Discovery’s VetScreenTM Cattle Sex determination kit was used for the determination of gender of meat samples in raw and cooked meat products. VIC-labelled probe target X chromosomal specific region, and FAM-labelled probes target X chromosomal specific region. Meat sample is reported as female origin if amplification is observed only in VIC channel, while FAM and VIC both channel got amplified in a case of meat sample obtained from a male species. (Figure 2). The results of all 15 samples for sex identification are given in Table 4. Only one buffalo meat sample was of female origin rest of the meat sample were of male origin.
Figure 2:
| Table 4: Gender determination. | |||
| Sample No | Biological Set Name | ♂Cq Fam | ♀Cq Vic |
| Sample 1 | buff | 31.22 | 33.76 |
| Sample 2 | buff | 35.41 | 34.15 |
| Sample 6 | cow | 30.02 | 28.68 |
| Sample 7 | cow | 32.17 | 30.62 |
| Sample 8 | cow | 36.76 | 33.32 |
| Sample 9 | cow | 35.27 | 29.81 |
| Sample 11 | cow | 39.08 | 29.63 |
| Sample 13 | cow | 33.99 | 33.79 |
| Sample 14 | cow | 35.12 | 33.46 |
| Sample 15 | cow | 34.39 | 33.77 |
| Sample 4 | Mix | 33.48 | 29.78 |
| Sample 5 | Mix | 33.21 | 32.74 |
| Sample 10 | Mix | 32.74 | 31.39 |
Authentication of meat assumes significance in view of religious, quality assurance, food safety, public health, conservation, and legal concerns [3]. Food labeling regulations require that the species of meat in food products are accurately declared to the consumer [28]. In a county like India due to religious sentiments as well as animal protection lows authentication of meat assumes significance hence, it is an important task for food control laboratories to be able to carry out species differentiation of raw materials to be used for industrial food preparation and the detection of animal species in food products [29]. The legal authorities need to find reliable ways to analyze the products and control the manufacturers by laws. Both the control mechanisms of the government and the producers need simple, low-cost, and applicable methods in order to sustain auto-control. Meat and meat products are very susceptible to spoilage and also expensive as compared to other food types. Therefore, their immediate, swift and reliable identification has great commercial as well as religious value.
To date molecular techniques are considered as reliable method to identify meat types [30]. Real time PCR is a method of choice as it has advantages such as high sensitivity and rapid performance with high sample numbers. Mitochondrial gene, conserved through evaluation is used here as a target for discrimination of species. The real time PCR is a fast (less turn-around time) and sensitive (can detect up to 10 copies of target). Forensic investigations where precise and reproducible techniques are needed, DNA based examination procedure have greater advantages over conventional methods [31]. The experiment conducted in this study was completed in less than 4 hour, which suggests that the accurate and precise identification of an unknown suspected meat sample is possible in a day.
Authors acknowledge Ms. Shefali Desai for reviewing the manuscript and providing constructive criticism, Ms. Khushboo Mishra for technical assistance and Mr. Gaurav Bhatt for the stimulating discussions.
- O’Mahony PJ. Finding horse meat in beef products–a global problem. QJM. 2013; 106: 595–597.
- Malnekoff E. Cattle Smuggling from India to Bangladesh. Honors Theses. 2013; 2378. scholarworks.wmich.edu/honors-theses/
- Kumar D, Singh SP, Karabasanavar NS, Singh R, Umapathi V. Authentication of beef, carabeef, chevon, mutton and pork by a PCR-RFLP assay of mitochondrial cytb gene. J Food Sci Technol. 2014; 51: 3458–3463. PubMed: https://pubmed.ncbi.nlm.nih.gov/26396346/
- Mane BG, Mendiratta SK, Tiwari AK, Bhilegaokar KN. Detection of Adulteration of Meat and Meat Products with Buffalo Meat Employing Polymerase Chain Reaction Assay. Food Anal Methods. 2012; 5: 296–300.
- Haider N, Nabulsi I, Al-Safadi B. Identifification of meat species by PCR-RFLP of the mitochondrial COI gene. Meat Sci. 2012; 90: 490–493. PubMed: https://pubmed.ncbi.nlm.nih.gov/21996288/
- Lou XP, Zhang W, Zheng J, Xu H, Zhao F. Comparative study on morphology of human, swine, sheep and cattle muscle tissues and its forensic significance. Fa Yi Xue Za Zhi. 2016; 32: 250–253. PubMed: https://pubmed.ncbi.nlm.nih.gov/29188664/
- Haunshi S, Basumatary R, Girish PS, Doley S, Bardoloi RK, et al. Identifification of chicken, duck, pigeon and pig meat by species-specifific markers of mitochondrial origin. Meat Sci. 2009; 83: 454–459. PubMed: https://pubmed.ncbi.nlm.nih.gov/20416682/
- Ishida N, Sakurada M, Kusunoki H, Ueno Y. Development of a simultaneous identification method for 13 animal species using two multiplex real-time PCR assays and melting curve analysis. Leg Med. 2018; 30: 64-71. PubMed: https://pubmed.ncbi.nlm.nih.gov/29197713/
- Rastogi G, Dharne MS, Walujkar S, Kumar A, Patole MS, et al. Species identifification and authentication of tissues of animal origin using mitochondrial and nuclear markers. Meat Sci. 2007; 76: 666–674. PubMed: https://pubmed.ncbi.nlm.nih.gov/22061243/
- Lockley AK, Bardsley RG. DNA-based methods for food authentication. Trends in Food Science & Technol. 2000; 11: 67-77.
- Hsieh HM, Chiang HL, Tsai LC, Lai SY, Huang NE, et al. Cytochrome b gene for species identification of the conservation animals. Forensic Sci Int. 2001; 122: 7-18. PubMed: https://pubmed.ncbi.nlm.nih.gov/11587860/
- Ennis S, Gallagher TF. A PCR-based sex-determination assay in cattle based on the bovine AMELogenin locus. Animal Genetics. 1994; 25: 425-427. PubMed: https://pubmed.ncbi.nlm.nih.gov/7695123/
- Hsieh YH, Sheu SC, Bridgman RC. Development of a monoclonal antibody specific to cooked mammalian meats. J Food Prot. 1998; 61: 476- 481. PubMed: https://pubmed.ncbi.nlm.nih.gov/9709213/
- Skarpeid HJ, Kvaal K, Hildrum KI. Identification of animal species in ground meat mixtures by multivariate analysis of isoelectricfocusing protein profiles. Electrophoresis. 1998; 19: 3103-3109. PubMed: https://pubmed.ncbi.nlm.nih.gov/9932802/
- Ebbehøj KF, Thomsen PD. Species differentiation of heated products by DNA hybridization. Meat Sci. 1991; 30: 221-234. PubMed: https://pubmed.ncbi.nlm.nih.gov/22061971/
- Janssen FW, Hägele GH, Buntjer JB, Lenstra JA. Species identification in meat by using PCR-generated satellite probes. J Industr Microbiol Biotechnol. 1998; 21: 115-120.
- Wolf C, Rentsch J, Hubner P. PCR-RFLP analysis of mitochondrial DNA: a reliable method for species identification. J Agric Food Chem. 1999; 47: 1350–1355. PubMed: https://pubmed.ncbi.nlm.nih.gov/10563979/
- Girish PS, Anjaneyulu ASR, Viswas KN, Shivakumar BM, Anand M, et al. Meat species identifification by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) of mitochondrial 12S rRNA gene. Meat Sci. 2005l; 70: 107–112. PubMed: https://pubmed.ncbi.nlm.nih.gov/22063286/
- Maede D. A strategy for molecular species detection in meat and meat products by PCR-RFLP and DNA sequencing using mitochondrial and chromosomal genetic sequences. Eur Food Res Technol. 2006; 224: 209–217.
- Sebastio P, Zanelli P, Neri TM. Identification of anchovy (Engraulisencrasicholus L.) and gilt sardine (Sardinella aurita) by polymerase chain reaction, sequence of their mitochondrial cytochrome b gene, and restriction analysis of polymerase chain reaction products in semi preserves. J Agric Food Chem.2001; 49: 1194-1199. PubMed: https://pubmed.ncbi.nlm.nih.gov/11312834/
- Asensio L, González I, Fernández A, Céspedes A, Rodríguez MA, et al. Identification of Nile perch (Lates niloticus), grouper (Epinephelusguaza), and wreck fish (Polyprion americanus) by polymerase chain reaction-restriction fragment length polymorphism of a 12S rRNA gene fragment. J Food Protection. 2000; 63: 1248-1252.
- Meyer R, Hoefelein C, Luethy J, Candrian U. Polymerase chain reaction-restriction fragment length polymorphism analysis: a simple method for species identification in food. Detection of soya in processed meat products. Z Lebensm Unters Forch. 1995; 203: 339–344.
- Jonker K, Tilburg J, Hagele G, de Boer E. Species identification in meat products using real-time PCR. Food Additives and Contaminants. 2008; 25: 527-533.
- Lopparelli RM, Cardazzo B, Balzan S, Giaccone V, Novelli E. Real-time TaqMan polymerase chain reaction detection and quantification of cow DNA in pure water buffalo mozzarella cheese: method validation and its application on commercial samples. J Agric Food Chem. 2007; 55: 3429-3434. PubMed: https://pubmed.ncbi.nlm.nih.gov/17419643/
- Aasen E, Medrano J. Amplification of the Zfy and Zfx Genes for Sex Identification in Humans, Cattle, Sheep and Goats. Nat Biotechnol. 1990; 8: 1279–1281. PubMed: https://pubmed.ncbi.nlm.nih.gov/1369448/
- Shi L, Yue W, Ren Y, Lei F, Zhao J. Sex determination in goat by amplification of the HMG box using duplex PCR. Animal Reproduction Science. 2008; 105: 398–403. PubMed: https://pubmed.ncbi.nlm.nih.gov/18096334/
- Anon. 2002.
- Ong SB, Zuraini MI, Jurin WG, Cheah YK, Tunung R, et al. Meat molecular detection: sensitivity of polymerase chain reaction-restriction fragment length polymorphism in species differentiation of meat from animal origin. ASEAN Food J. 2007; 14: 51–59.
- Luo J, Wang J, Bu D, Li D, Wang L, et al. Development and application of a PCR approach for detection of beef, sheep, pig, and chicken derived materials in feedstuff. Agr Sci China. 2008; 7: 1260–1266.
- Kumar A, Kumar RR, Sharma B, Gokulakrishnan P, Mendiratta SH, et al. Identification of Species Origin of Meat and Meat Products on the DNA Basis: A Review, Critical Reviews in Food Science and Nutrition. 2015; 55: 1340-1351. PubMed: https://pubmed.ncbi.nlm.nih.gov/24915324/
- Bellisa C, Ashtona KJ, Freneyb L, Blairb B, Griffifiths LR. A molecular genetic approach for forensic animal species identification. Forensic Sci Int. 2003; 134: 99–108. PubMed: https://pubmed.ncbi.nlm.nih.gov/12850402/